
Robust
Uniform coverage for accurate CNV results.

Reliable
High gene coverage for comprehensive CNV detection.

Simple
Six easy steps with minimal hands on time and direct-to-sequencer loading capability.
Our digitalMLPA™ products are for research use only (RUO). Not for use in diagnostic procedures.
digitalMLPA: unparalleled broad copy number certainty
digitalMLPA™ is a powerful combination of our trusted SALSA® MLPA® technology and next-generation sequencing, meeting your large-scale copy number detection needs. With digitalMLPA, you can analyse a greater number of targets in a single reaction with only 20 ng of input DNA, while also reducing costs and turnaround times.

Applications
Our current panel range covers hereditary cancer markers and hematological malignancies such as multiple myeloma and acute lymphoblastic leukaemia. The digitalMLPA portfolio also features an assay for genome-wide detection of large-scale copy number changes. Additional assays are in development for an even broader range of applications.
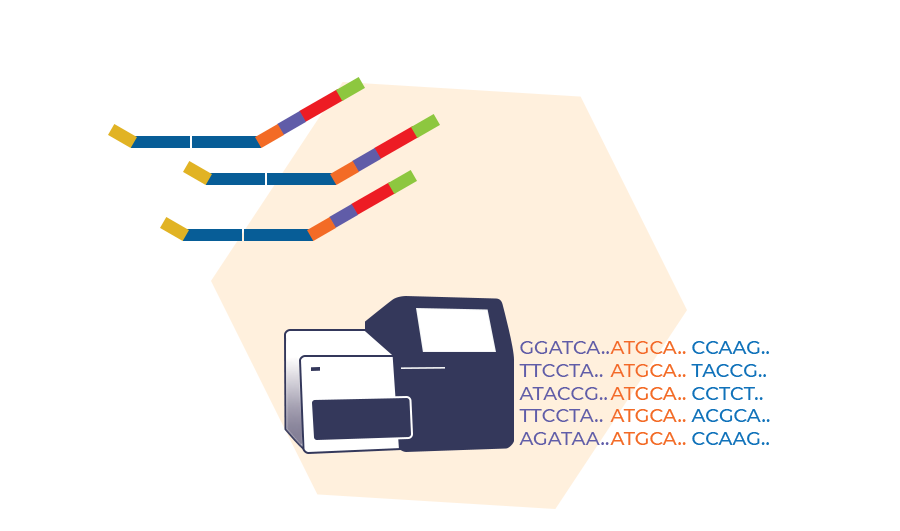
How does digitalMLPA work?
digitalMLPA is a PCR-based technique that enables the detection of up to 1200 targeted DNA sequences in a single multiplex reaction. After the PCR-based amplicon generation, each probe amplicon is quantified using an Illumina NGS platform. digitalMLPA does not require post-PCR clean up or library concentration measurements, making the technique efficient and user-friendly.
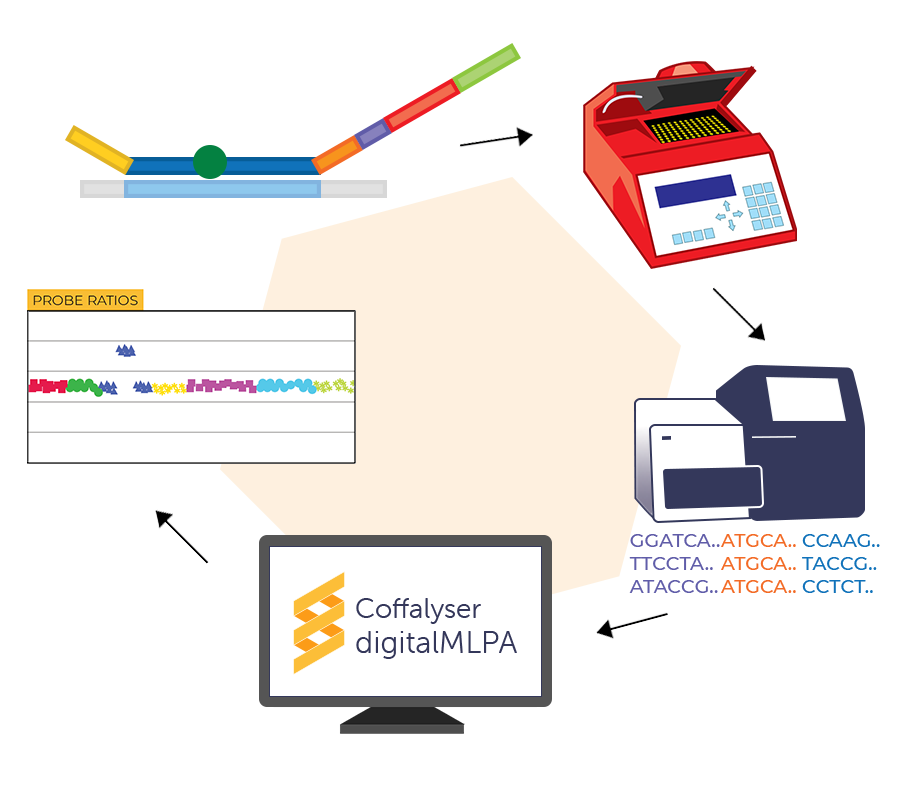
Coffalyser digitalMLPA
Coffalyser digitalMLPA™ is our free and easy-to-use data analysis software that makes analysing digitalMLPA experiments fast and effortless. Coffalyser digitalMLPA can automatically recognise digitalMLPA sequence reads from FASTQ files, and directly uses them for data analysis. Thanks to its advanced quality checks and algorithms, Coffalyser digitalMLPA can quickly detect and analyse aberrations in your samples, providing you with clear and detailed reports for every experiment.

Can We Help You?
Contact our support team.



